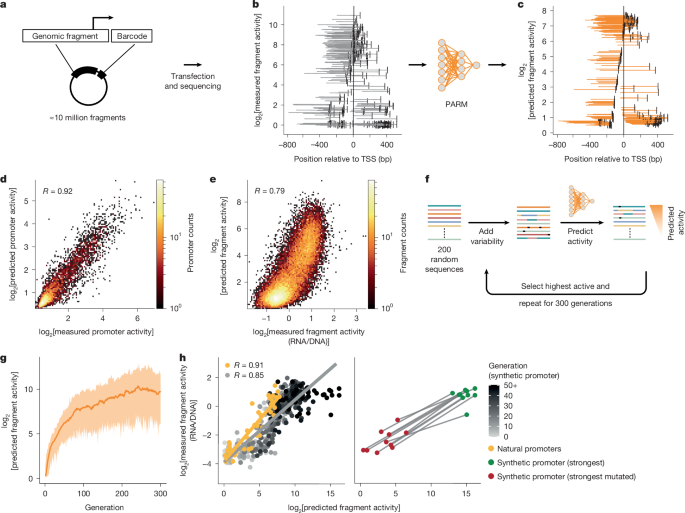
"Promoters generally consist of a transcription start site (TSS) and several hundreds of base pairs of DNA (mostly upstream of the TSS) that contain short sequence motifs that can be bound by a multitude of transcription factors (TFs)2,3. The construction of computational models that can predict promoter activity from a DNA sequence is challenging. Deep-learning techniques hold promise for this purpose1,4,5,6 but depend on substantial training datasets. In one approach, this data mass is obtained by aggregating hundreds or thousands of genome-wide maps of transcription and epigenome features from many cell types7,8,9,10,11,12,13."
"MPRAs offer an alternative source of training data that can be obtained for a single specific cell type. Here millions of genomic DNA sequences, each several hundreds of base pairs long, are tested for their autonomous regulatory activity in the cell type of interest. Because the fragments are tested in isolation, measured activity can be unambiguously assigned; therefore, inference of the causal roles of specific DNA sequences may be more straightforward than with epigenome and transcriptome maps (Fig. 1a)."
Promoters comprise a transcription start site (TSS) and several hundred base pairs of upstream DNA containing short sequence motifs bound by transcription factors. Predicting promoter activity from DNA sequence is challenging. Deep learning can help but requires substantial training datasets often built by aggregating hundreds or thousands of genome-wide transcription and epigenome maps across many cell types, which demands extensive computational resources. Epigenomic profiles are imperfect proxies, can be confounded by long-range autocorrelation and are only correlative, complicating causal inference. Some models revealed promoter grammar features but cannot predict regulation in unseen cell types. Massively parallel reporter assays provide cell-type-specific, isolated tests of millions of sequences, allowing unambiguous activity assignment and more direct causal inference.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]