"Bulk RNA-sequencing studies of ageing have revealed disruptions to essential cellular processes such as transcription, translation and growth-factor signalling3, with processes involved in mitochondrial function, neuronal activity and DNA damage being dysregulated in the ageing brain2,4. Cell-type-specific changes during ageing are obscured in bulk analyses and are poorly understood. This represents a major knowledge gap in the human brain, in which molecularly distinct cell types perform specific functions throughout life."
"The advent of single-cell genomics has allowed high-resolution analysis of both DNA and RNA. scWGS and other techniques have shown that somatic mutations accumulate in human neurons during ageing and in age-related diseases, raising the possibility that such variants contribute to transcriptional dysregulation and the concomitant increased susceptibility to dysfunction and disease that accompanies advanced age5,6,7,8,9,10. Single-cell RNA sequencing and snRNA-seq have refined the understanding of brain cell states11,12,13,14."
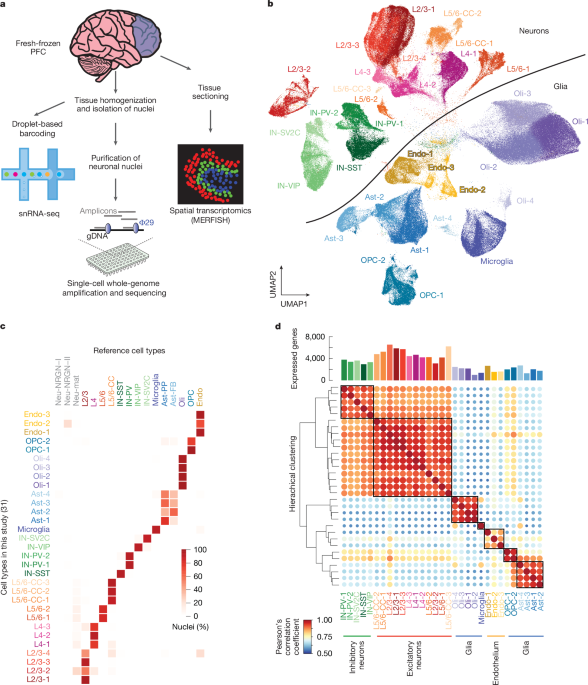
Bulk RNA-sequencing of ageing reveals disruptions to transcription, translation, and growth-factor signalling, with mitochondrial function, neuronal activity, and DNA damage pathways dysregulated in the ageing brain. Cell-type-specific changes during ageing are obscured in bulk analyses and remain poorly understood, creating a major knowledge gap given the functional diversity of molecularly distinct brain cell types. Single-cell genomics shows somatic mutations accumulate in human neurons during ageing and age-related diseases, suggesting contributions to transcriptional dysregulation and increased susceptibility to dysfunction. Droplet-based snRNA-seq and scWGS were applied to fresh-frozen human prefrontal cortex across a lifespan of neurotypical donors, with MERFISH used for spatial-transcriptomic validation.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]