"Mammalian genomes are extensive repositories of DNA-encoded instructions that control cellular functions through complex regulatory mechanisms. Central to this regulation are cis-regulatory elements (CREs)-non-coding DNA sequences that control the transcription of nearby genes1. Usually associated with open chromatin and specific histone modifications, CREs contain binding sites for transcription factors and other chromatin-associated proteins that interact with one another and the transcriptional machinery to regulate gene expression."
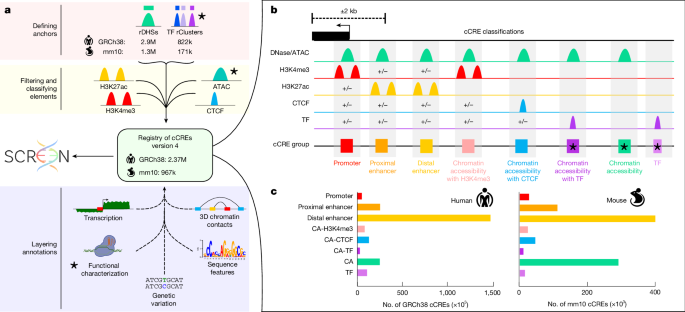
"Here we have expanded the registry of cCREs to include 2.37 million human and 967,000 mouse elements. This expansion leverages new datasets produced during ENCODE Phase IV (ENCODE4) and improved computational methods, making the registry one of the most extensive repositories of CREs available. The updated registry spans hundreds of unique cell and tissue types, and enhances our understanding of gene regulation across a broad range of biological contexts."
The registry of candidate cis-regulatory elements (cCREs) expanded to include 2.37 million human and 967,000 mouse elements. The expansion uses new ENCODE Phase IV datasets and improved computational methods to create one of the most extensive CRE repositories. The updated registry covers hundreds of unique cell and tissue types, enabling broader characterization of gene regulation across biological contexts. Functional characterization data were integrated for more than 97% of human cCREs, revealing relationships between sequence features and regulatory activity and exposing new functional subclasses. Thousands of silencer cCREs were identified, many of which function as enhancers in alternative cellular contexts.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]