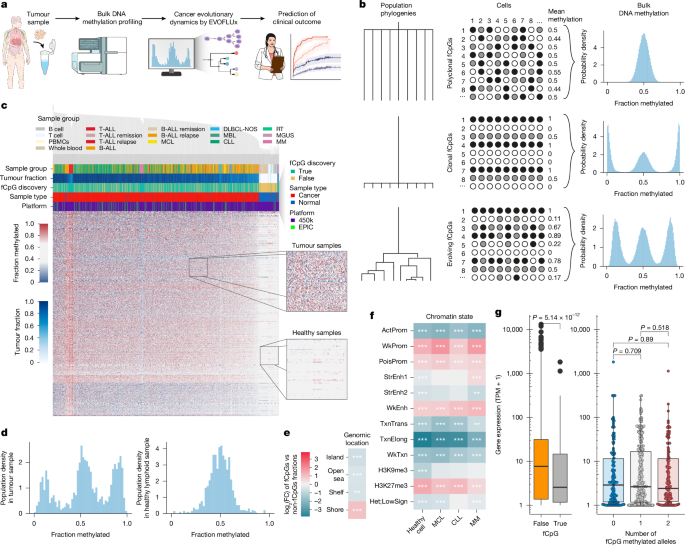
"DNA methylation can serve as a lineage marker, recording the clonal architecture of cell populations9,10,11,12,13 or the proliferative history14,15. We have recently identified DNA methylation at some CpG sites, which stochastically fluctuates over time at a timescale measured in years4. These fluctuating CpGs (fCpGs) function as a 'methylation barcode', providing a low-cost strategy to provide high temporal resolution lineage tracing in patient samples4."
"Cancer development is an evolutionary process1,2; consequently, the evolutionary history of a cancer may set its future trajectory and allow inference of the clinical path of a patient5. However, testing this hypothesis directly is challenging because longitudinal patient samples are required to document evolutionary history. Consequently, evolutionary histories are typically inferred from single timepoint data; for example, somatic (epi)mutations are patterned in distinctive ways by differing evolutionary dynamics3."
Cancer development is an evolutionary process, and the evolutionary history of a cancer can set its future trajectory and clinical path. Longitudinal patient samples are required to document evolutionary history, making direct testing challenging. Single-cell genome sequencing infers phylogenies but is expensive and limited in case numbers. DNA methylation at some CpG sites fluctuates stochastically over years and can record clonal architecture and proliferative history. Fluctuating CpGs (fCpGs) act as a methylation barcode enabling low-cost, high-temporal-resolution lineage tracing. EVOFLUx is a quantitative modelling framework that infers evolutionary histories from fCpG data from clinical specimens at scale. Bulk cancer methylation arrays enable inferring dynamics for correlation with clinical variables and outcomes.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]