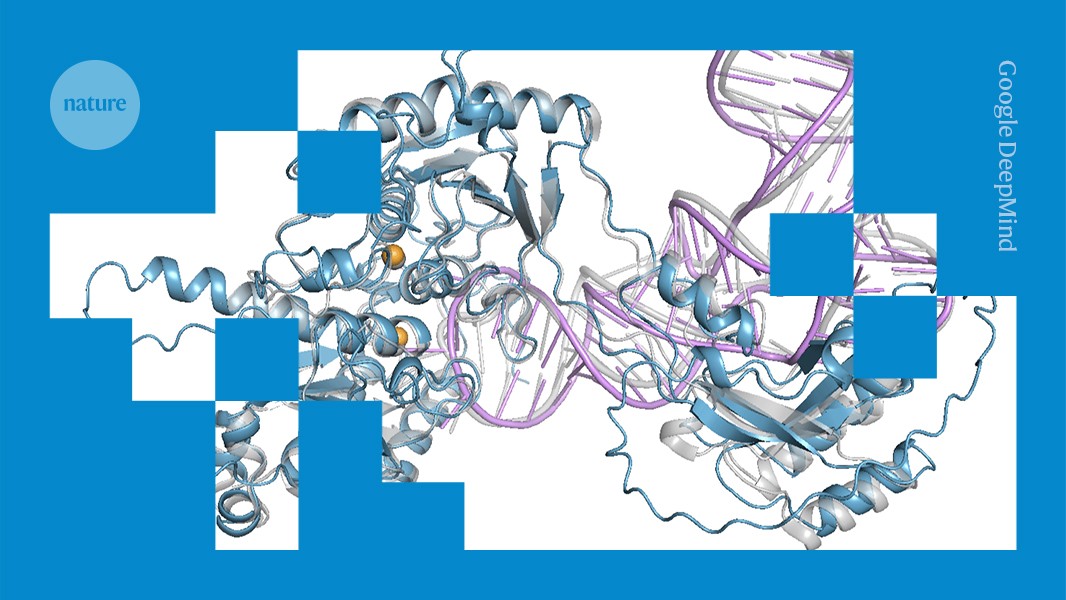
"The tool still doesn't have the same functionality as AlphaFold3, but "we wanted to get something out to the community as soon as possible", says Woody Sherman, the consortium's executive committee chair and chief innovation officer at Psivant Therapeutics in Boston, Massachusetts. The team hopes to use researcher feedback after the preview release to improve the model. Full release coming The system was trained on more than 300,000 molecular structures and a synthetic database of more than 40 million structures."
"Unlike AlphaFold3, which is available for restricted academic use, any researcher or pharmaceutical company can use OpenFold3. "It's a big step forward in terms of the democratization of AI structural-biology tools," says Sherman. The preview release shares OpenFold3's code with researchers and allows them to start using and testing it. Sherman says that the consortium team is still working on technical improvements to the system and is planning a full release in the coming months."
OpenFold3 is an open-source AI system developed by the OpenFold Consortium to predict protein 3D structures from amino-acid sequences and to model molecular interactions with drugs or DNA. The system was trained on more than 300,000 experimental structures plus a synthetic database exceeding 40 million structures, and development has cost about US$17 million so far. The preview release shares code widely so researchers and companies can use and test the model. The system is not yet fully equivalent to AlphaFold3; the team seeks researcher feedback, plans technical improvements, and expects a full release in coming months.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]