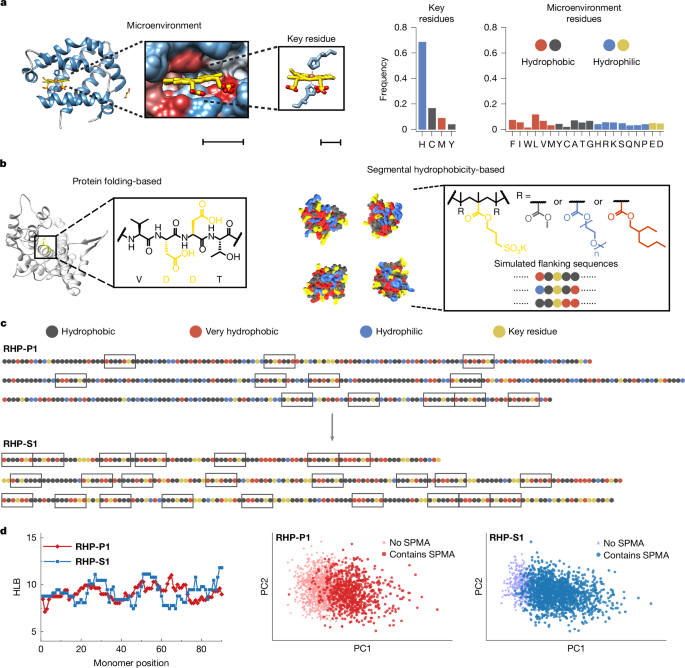
"We propose that for polymers with backbone chemistries different from that of proteins, programming spatial and temporal projections of sidechains at the segmental level can be effective in replicating protein behaviours13,14; and leveraging the rotational freedom of polymer can mitigate deficiencies in monomeric sequence specificity and achieve behaviour uniformity at the ensemble level2,3,15,16,17,18,19,20. Here, guided by the active site analysis of about 1,300 metalloproteins, we design random heteropolymers (RHPs) as enzyme mimics based on one-pot synthesis."
"We introduce key monomers as the equivalents of the functional residues of protein and statistically modulate the chemical characteristics of key monomer-containing segments, such as segmental hydrophobicity21. The resultant RHPs form pseudo-active sites that provide key monomers with protein-like microenvironments, co-localize substrates with catalytic or cofactor-binding sidechains and catalyse reactions such as oxidation and cyclization of citronellal with isopulegol/menthoglycol selectivity. This RHP design led to enzyme-like materials that can retain catalytic activity under non-biological conditions, are compatible"
Design of random heteropolymers leverages programmed spatial and temporal sidechain projections at the segmental level to emulate protein functions despite differing backbone chemistries. Active site analysis of roughly 1,300 metalloproteins informed monomer selection and statistical modulation of segment properties such as hydrophobicity. One-pot synthesis produces RHPs that form pseudo-active sites, creating protein-like microenvironments that co-localize substrates with catalytic or cofactor-binding sidechains. These materials catalyse oxidation and cyclization reactions, achieving isopulegol/menthoglycol selectivity for citronellal. The resulting enzyme-like polymers retain catalytic activity under non-biological conditions.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]