"Although generative artificial intelligence (AI) promises to accelerate the design of functional biological systems, articulating 'function' to a generative model remains challenging and often underspecified. In natural language, distributional semantics hypothesizes that meaning can be represented by word co-occurrence, that is, 'you shall know a word by the company it keeps'3,4 (Fig. 1a). In biology, an emerging distributional hypothesis defines the function of a gene through its interactions with other genes, that is, 'you shall know a gene by the company it keeps'2."
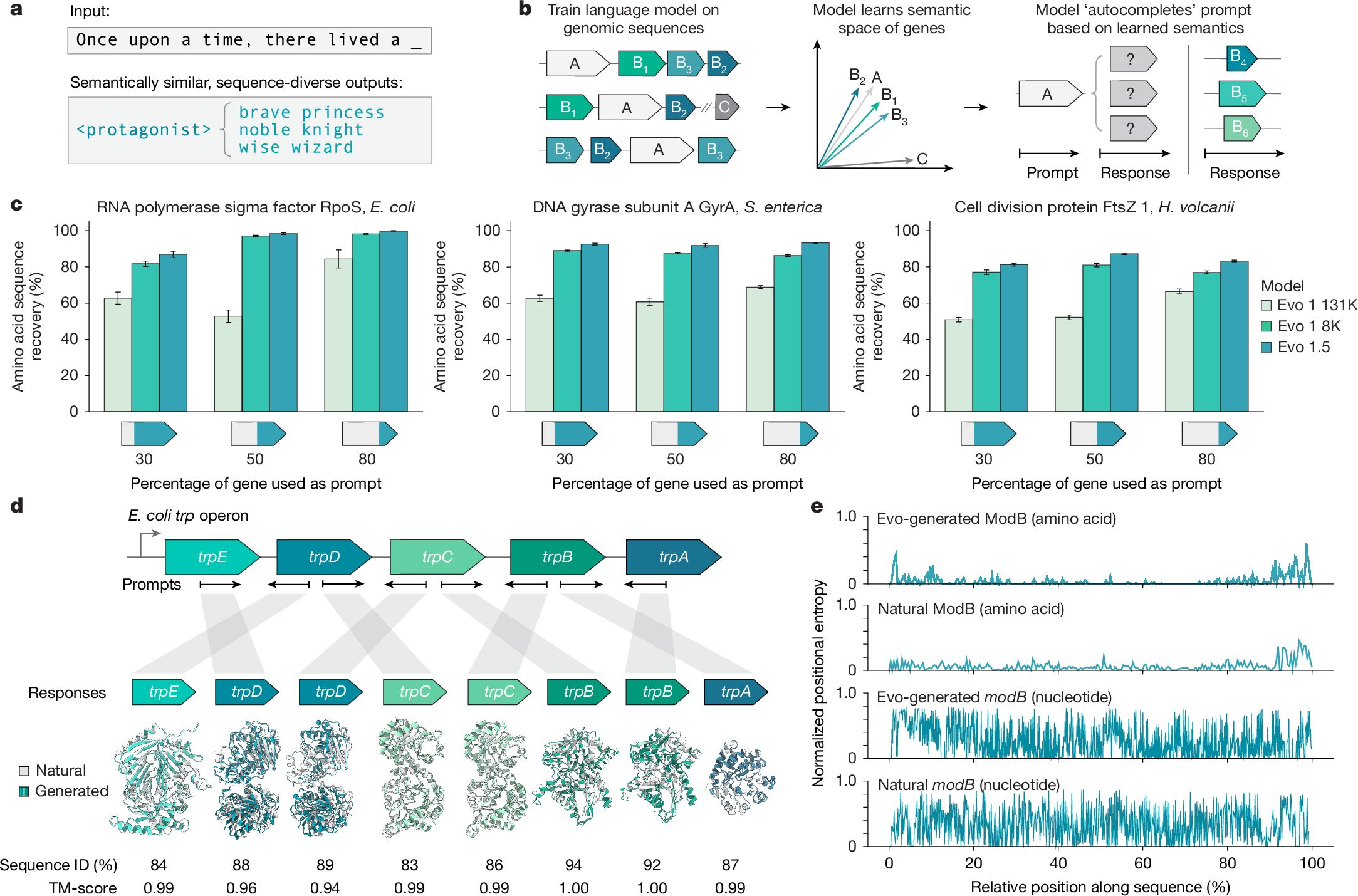
"b, In semantic design, a genomic language model trained across multiple genes learns to map genes with related functions to similar semantic spaces, enabling the generation of functionally related yet sequence-diverse genes. c, Sequence recovery assessments, where a genomic language model is used to autocomplete three conserved prokaryotic genes, show consistent improvements from Evo 1 131K and Evo 1 8K to Evo 1.5, reflecting an enhanced ability to leverage genomic context."
Distributional semantics links meaning to word co-occurrence, and a genomic analogue defines gene function by interactions with neighbouring genes. A genomic language model trained across multiple genes maps functionally related genes to similar semantic spaces, enabling generation of functionally related yet sequence-diverse genes. Sequence recovery assessments on three conserved prokaryotic genes show consistent improvements across model versions, indicating enhanced leverage of genomic context. Completion of conserved E. coli trp operon genes from both strands yields high sequence recovery and predicted structural conservation. Positional entropy comparisons reveal conserved essential amino acids alongside high nucleotide diversity.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]