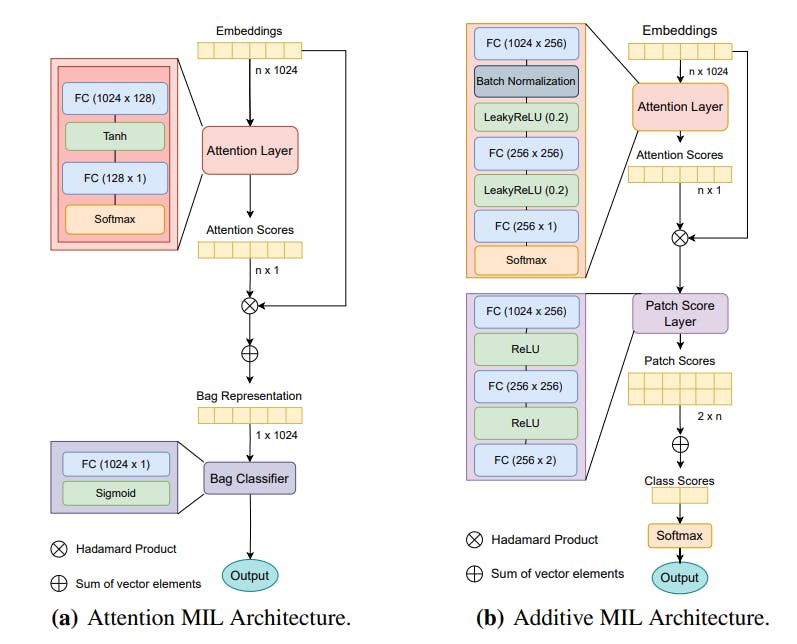
"Whole Slide Images (WSI) are essential in Digital Pathology, yet they pose challenges for AI analysis as labeling is usually at the slide level, not tile level."
"To tackle the detection of oncogene mutations, a weakly supervised Multiple Instance Learning (MIL) approach was investigated for Invasive Breast Carcinoma and Lung Squamous Cell Carcinoma."
"The results indicated that a novel additive implementation of MIL performs comparably to reference methods in tumor detection and identifying TP53 mutations."
"The dual challenge lies in accurately predicting the overall cancer phenotype while pinpointing associated cellular morphologies at the tile level."
Whole Slide Images (WSI) are fundamental to Digital Pathology, yet they create challenges for AI analysis because pathology labeling occurs at the slide level rather than the tile level. This challenge encompasses both the accurate prediction of cancer phenotypes and understanding the cellular morphologies related to them at the tile level. A weakly supervised Multiple Instance Learning (MIL) technique was applied to Invasive Breast Carcinoma and Lung Squamous Cell Carcinoma to aid in tumor detection and TP53 mutations, ultimately showcasing a novel implementation of MIL that achieves performance on par with established methods.
Read at Hackernoon
Unable to calculate read time
Collection
[
|
...
]