"In the past decade, increasingly sensitive sequencing methods have begun to unravel the somatic mutation landscapes of human tissues. They have revealed that mutations accumulate linearly with age in a tissue-specific manner, largely due to endogenous mutational processes but also influenced by mutagen exposures, germline variation and disease states. These studies have also revealed that as we age our tissues are colonized by myriad clones carrying positively selected driver mutations1,2,3,4,5,6,7. These clones provide a window into early carcinogenesis and may contribute to other diseases."
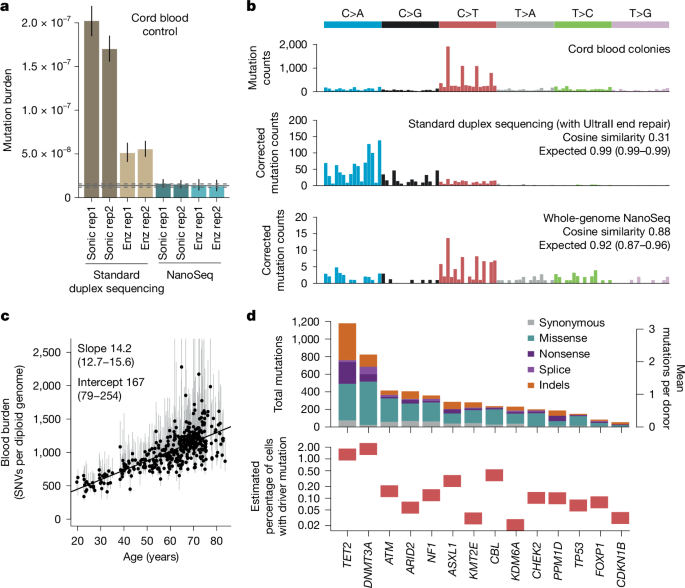
"We have previously described NanoSeq, a protocol that avoids error transfer by using restriction enzyme fragmentation without end repair, and dideoxynucleotides during A-tailing, achieving error rates below 5 × 10 −9 errors per bp in single DNA molecules8. As this rate is two orders of magnitude lower than the mutation burden of normal adult cells (around 10 −7), mutations are accurately detected from single DNA molecules, enabling the quantification of mutation rates and signatures in any tissue."
Sensitive sequencing has revealed that somatic mutations accumulate linearly with age in a tissue-specific manner, driven largely by endogenous mutational processes and modulated by mutagen exposures, germline variation and disease states. Aging tissues become colonized by numerous microscopic clones carrying positively selected driver mutations, which offer insight into early carcinogenesis and may contribute to other diseases. Most clones are microscopic and current detection methods are low throughput, limiting cohort and tissue coverage. Error-corrected bulk approaches like duplex sequencing reduce errors, while NanoSeq minimizes interstrand error transfer to achieve error rates below 5 × 10 −9 per bp, enabling accurate single-molecule mutation detection but lacking full-genome coverage for driver discovery.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]