"For more than a decade, innovation in high-throughput DNA sequencing (that is, transforming information stored in DNA into human- and machine-readable sequences) has propelled research in the biomedical domain and led to an exponential growth in worldwide sequencing capacity14,15. A large proportion of these data is deposited in publicly funded repositories, such as the European Nucleotide Archive (ENA) maintained by the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI)16,"
"Although the rate of data deposition has slowed in recent years, the number of sequenced nucleotides contained in the ENA still doubles roughly every 45 months and currently comprises more than 10.8 × 10 16 nucleotide bases (108 Pbp)18 of raw sequencing data, of which 6.7 × 10 16 nucleotide bases (67 Pbp) are publicly available (as of 11 January 2025; Supplementary Fig. 13). The classical pattern for accessing sequencing data in such large repositories is to identify relevant records using descriptive metadata and to retrieve a copy or slice of the data for further processing."
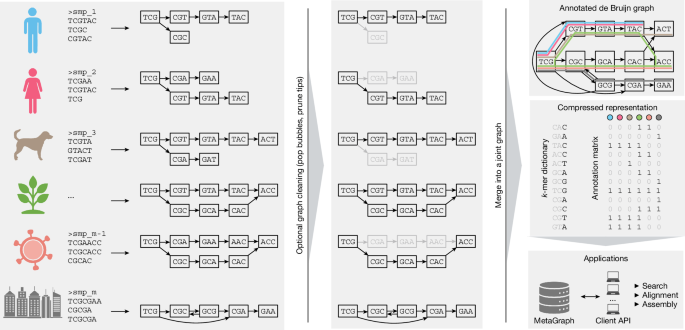
High-throughput DNA sequencing has driven exponential growth in global sequencing capacity over more than a decade. Public repositories such as the European Nucleotide Archive, the Sequence Read Archive, and the DDBJ Sequence Read Archive store vast volumes of raw sequencing data. The ENA contains over 10.8×10^16 nucleotide bases of raw data, with 6.7×10^16 bases publicly available as of 11 January 2025. Traditional access relies on descriptive metadata queries and downloading or cloud access, which requires considerable resources. Petabase-scale raw sequencing data remained inaccessible for full-text sequence search, limiting research utility. A highly scalable indexing approach converts these repositories into compressed, portable representations suitable for full-text search and downstream analysis.
#high-throughput-sequencing #sequence-data-repositories #petabase-scale-indexing #compressed-sequence-representations
Read at Nature
Unable to calculate read time
Collection
[
|
...
]